NASA’s Juno Spacecraft Completes Flyby Over Jupiter’s Great Red Spot



NASA’s Juno Spacecraft Completes Flyby over Jupiter’s Great Red Spot
NASA’s Juno mission completed a close flyby of Jupiter and its Great Red Spot on July 10, during its sixth science orbit.
All of Juno’s science instruments and the spacecraft’s JunoCam were operating during the flyby, collecting data that are now being returned to Earth. Juno’s next close flyby of Jupiter will occur on Sept. 1.
Raw images from the spacecraft’s latest flyby will be posted in coming days.
“For generations people from all over the world and all walks of life have marveled over the Great Red Spot,” said Scott Bolton, principal investigator of Juno from the Southwest Research Institute in San Antonio. “Now we are finally going to see what this storm looks like up close and personal.”
The Great Red Spot is a 10,000-mile-wide (16,000-kilometer-wide) storm that has been monitored since 1830 and has possibly existed for more than 350 years. In modern times, the Great Red Spot has appeared to be shrinking.
Juno reached perijove (the point at which an orbit comes closest to Jupiter’s center) on July 10 at 6:55 p.m. PDT (9:55 p.m. EDT). At the time of perijove, Juno was about 2,200 miles (3,500 kilometers) above the planet’s cloud tops. Eleven minutes and 33 seconds later, Juno had covered another 24,713 miles (39,771 kilometers), and was passing directly above the coiling crimson cloud tops of the Great Red Spot.
The spacecraft passed about 5,600 miles (9,000 kilometers) above the clouds of this iconic feature.
On July 4 at 7:30 p.m. PDT (10:30 p.m. EDT), Juno logged exactly one year in Jupiter orbit, marking 71 million miles (114.5 million kilometers) of travel around the giant planet.
Juno launched on Aug. 5, 2011, from Cape Canaveral, Florida. During its mission of exploration, Juno soars low over the planet’s cloud tops – as close as about 2,100 miles (3,400 kilometers). During these flybys, Juno is probing beneath the obscuring cloud cover of Jupiter and studying its auroras to learn more about the planet’s origins, structure, atmosphere and magnetosphere.
Early science results from NASA’s Juno mission portray the largest planet in our solar system as a turbulent world, with an intriguingly complex interior structure, energetic polar aurora, and huge polar cyclones.
JPL manages the Juno mission for the principal investigator, Scott Bolton, of Southwest Research Institute. The Juno mission is part of the New Frontiers Program managed by NASA’s Marshall Space Flight Center in Huntsville, Alabama, for the Science Mission Directorate.
Lockheed Martin Space Systems, Denver, built the spacecraft. JPL is a division of Caltech in Pasadena
More Posts from Contradictiontonature and Others

We might think we know the human body pretty well by now, but scientists are still discovering incredible individuals who are defying all odds by living out their lives with crucial parts missing, added, or tweaked in the most extraordinary ways.
From those with almost superhuman abilities, to others living without the organs we hold most dear, here are five of the most remarkable humans known to medicine.
Read more…
Checking Cancer At Its Origin..
In a first, the lab led by Leonard Zon at Boston Children’s Hospital has visualised the emergence of the primary melanoma cell in transgenic zebrafish that harbour the human oncogenic BRAFV600E mutation in melanocytes. This cancerous state is characterised in maturing fish by the formation of neural crest progenitors [NCPs], which are the predecessors of melanocytes and are only seen in the embryonic stage of healthy zebrafish.
The Zon lab placed the human mutated oncogene, BRAFV600E (a characteristic of benign human nevi/moles) under the control of a melanocyte-specific promoter and introduced it into the zebrafish. Generations of this transgenic fish were engineered such that they were also deficient in functional p53 (loss of function mutation). They used previous findings that in healthy zebrafish, a gene called crestin is expressed only in the embryonic NCPs and never throughout maturity, but is re-expressed selectively in melanomatous cells during adulthood. crestin was cloned adjacent to a reporter, enhanced green fluorescent protein [EGFP] for live imaging purposes.
The developmental phases of the fish, that were by now triple transgenic (for human BRAFV600E, p53 LOF and crestin:EGFP) were observed by live imaging; ~21 days after fertilisation, the expression of crestin:EGFP localised precisely to the (future) melanoma sites, and the very first triple-transgenic (individual) cells that went on to form larger masses of cells were also observed. To summarise, melanoma formation was observed in three stages: individual fluorescent cells, followed by these cells multiplying to form groups of <50 cells, and lastly these groups forming raised lesions. This consistently held true, with all 30 observed individual cells turning into 30 lesions. These results are illustrated in Figure 1.

Figure 1. In the top left box, a single cell is visualised as it multiplies into a group of melanoma cells (top right). The bottom images show the raised melanoma lesion as observed by the naked eye and by live imaging. The green fluorescence emitted from EGFP indicates that it is localised only to the melanoma (as is crestin expression), that is, it has not metastasised elsewhere.
These pre-cancerous cells were also shown to be self-sustaining and tumourigenic: when fish scales containing the mutant cells were transplanted to another part of the same fish (auto-transplant) or to another fish (allo-transplant) that was also exposed to radiation, the cells proliferated in the new site, as well as penetrated the hypodermis underneath (Figure 2).

Figure 2. The fluorescence indicates a single scale being auto-transplanted elsewhere on the same fish. As the days progress, the patch expands as well, and after day 33, the cells penetrate deeper into the hypodermis and thrive independently, and excising the transplanted scale proves futile.
Role of Transcription Factor sox10
sox10 is a master TF in NCP and its over-expression has been correlated with increased crestin expression, and accordingly, sox10 over expression in the transgenic melanocytes accelerated the melanoma onset. Following the logical train of thought that sox10 promotes melanoma progression, it was then targeted by CRISPR-Cas9 and inactivated in the transgenic cells. This resulted in a delayed onset of melanoma (180 days) compared to the controls (133 days). sox10 is also known to be expressed in most human melanoma cell lines. Moreover, the DNA element that acts as the binding site for Sox10 is also found in a hyper-acetylated [H3K27Ac], super-enhancer state. This is an epigenetic alteration and may prove a useful target in therapy (ex. HAT inhibitors).
Summary
The key finding clears up a hitherto ambiguous association between a reversion to stem/progenitor cell-like status and cancer: it indicates that the apparent devolution of a specialised cell to a primitive cellular state is not a consequence of cancer progression, but that it is an hallmark of pre-cancerous cells that may contribute to tumour progression. The rarity of melanoma formation among the mutant cells also suggests that the double mutant [BRAFV600E; p53 LOF] is not the only factor to influence the onset. Experimentally, crestin expression was a definitive prelude to formation of nevi which transformed into full-fledged raised melanomas in that spot.
This discovery has two chronological applications: first, of the many susceptible melanocytes harbouring the mutated oncogene, we can find out which are most likely to enter the melanoma state. Peaks in the expression profile of sox2, or a couple other TFs, dlx2 and tfap2, can prove to be a telltale pre-melanoma signature and thus be used in diagnosis. Secondly, by doing so, these can be better targeted early on before they’ve disseminated and become virtually untreatable.
Kaufman CK, Mosimann C, Fan ZP, Yang S, Thomas AJ, Ablain J, et al. A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science. 2016;351[6272]:aad2197–aad2197.










TOP TEN MOST DEADLY INFECTIOUS DISEASES
This list is based off of the assumption that the infected individual does not receive medical treatment.
1. Prions (mad cow disease, Creutzfeld-Jakob disease, kuru, fatal familial insomnia): 100%
2. Rabies: ~100%
3. African trypanosomiasis (’African sleeping sickness’): ~100%
4. Primary amoebic encephalitis caused by Naegleri fowlerii (’the brain-eating amoeba’): ~100%
5. Yersinia pestis, specifically the pneumonic or septicemic subtype (’the black plague’): ~100%
6. Visceral leishmaniasis: ~100%
7. Smallpox, specifically the malignant (flat) or hemorragic subtype: 95%
8. Ebola virus, specifically the Zaire strain: 83-90%
9. HIV: 80-90%
10. Anthrax, specifically the pulmonary subtype: >85%
One of the smoothest, most beautiful color changes I’ve ever seen.
The reaction is methoxymethyl deprotection of one of my agonists with concentrated HCl in acetonitrile as my solvent. The color change doesn’t happen in THF!

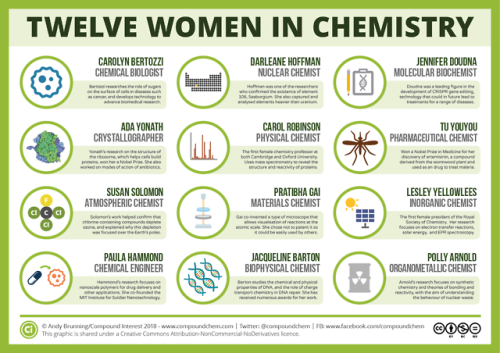
For International Women’s Day, here are 12 women from chemistry history: wp.me/p4aPLT-2ra and 12 from chemistry present: wp.me/p4aPLT-5w7










Watch: Bill Nye uses science to defend women’s reproductive rights
Follow @the-future-now

Viruses support photosynthesis in bacteria: An evolutionary advantage?
Viruses propagate by infecting a host cell and reproducing inside. This not only affects humans and animals, but bacteria as well. This type of virus is called bacteriophage. They carry so called auxiliary metabolic genes in their genome, which are responsible for producing certain proteins that give the virus an advantage. Researchers at the University of Kaiserslautern and the Ruhr University Bochum have analysed the structure of such a protein more closely. It appears to stimulate the photosynthesis of host bacteria. The study has now been published in the journal The Journal of Biological Chemistry.
Raphael Gasper, Julia Schwach, Jana Hartmann, Andrea Holtkamp, Jessica Wiethaus, Natascha Riedel, Eckhard Hofmann, Nicole Frankenberg-Dinkel. Auxiliary metabolic genes- Distinct Features of Cyanophage-encoded T-type Phycobiliprotein Lyase θCpeT. Journal of Biological Chemistry, 2017; jbc.M116.769703 DOI: 10.1074/jbc.M116.769703
The association between the virus protein and bacterial pigment is incredibly stable. Furthermore, the complex is highly fluorescent. Credit: AG Frankenberg-Dinkel
Complement Pathways

Components of complement pathways of the immune system.
Classical Pathway: binds to the pathogen surface
C1 binds to phosphocholine on bacteria, which activates C1r to cleave C1s.
Activated C1s cleaves C4 to C4a and C4b.
C4b binds to the microbial surface and also binds C2.
C2 is cleaved to C2a and C2b by C1s, forming the C4bC2a complex.
The C4bC2a complex cleaves C3 to C3a and C3b.
C3b binds to the surface and causes opsonization.
MB-Lectin Pathway: uses mannin-binding lectin to bind to mannose-containing carbohydrates on the pathogen surface
Mannin-binding lectin (MBL) binds to the pathogen surface and activates MASP-2.
MASP-2 cleaves C4 to C4a and C4b.
C4b binds to the microbial surface and also binds C2.
C2 is cleaved to C2a and C2b by MASP-2, forming the C4bC2a complex.
The C4bC2a complex cleaves C3 to C3a and C3b.
C3b binds to the surface and causes opsonization.
Alternative Pathway: binds to the pathogen surface with spontaneously activated complement, amplifies C3b
C3b deposited by the C3 convertase binds to factor B.
Factor B is cleaved by factor D into Ba and Bb, forming the C3bBb complex.
The C3bBb complex cleaves C3 into C3a and C3b.
C3 spontaneously hydrolyzes to C3(H2O).
C3(H2O) binds to factor B, and factor D cleaves factor B.
Upon factor B cleavage, the C3(H2O)Bb complex is formed.
The C3(H2O)Bb complex cleaves C3 into C3a and C3b.
Factor B binds to C3b on the surface and is cleaved to Bb.

Marrow Christmas and a Happy New Smear!
A very seasonal smear made from red marrow extracted from the iliac crest of a donor’s pelvis prior to transplantation.
Happy Holidays everyone
i♡histo
The image amazingly captures a single moment in time during the development of thousands of red and white blood cells.
Many of the small cells that are visible, like the ones forming the snowman’s carrot nose, do not have a nucleus. These are brand new erythrocytes (red blood cells) that are ready to exit the bone and enter the blood stream.
The other, slightly larger cells that have nuclei, like the snowman’s eyes and his top button, are either precursors to these erythrocytes (they will mature and lose their nucleus) or are precursors to the other blood cells in our body, the leukocytes (white blood cells): lymphocytes, monocytes, neutrophils, eosinophils and basophils.
In addition, the bone marrow is home to the cells that form platelets. These are huge multinucleated cells aptly named megakaryocytes - perhaps the cell at the bottom right.
It is possible to identify each mature cell and its precursor based upon its morphology and staining at higher magnification. High or low levels of these cells can indicate disease or cancers of the blood.
-
 hufflepuff-95 reblogged this · 5 years ago
hufflepuff-95 reblogged this · 5 years ago -
 sonicsoundscapes liked this · 6 years ago
sonicsoundscapes liked this · 6 years ago -
 inefable-enigma liked this · 7 years ago
inefable-enigma liked this · 7 years ago -
 julianajohnsonphoto liked this · 7 years ago
julianajohnsonphoto liked this · 7 years ago -
 mojomatao reblogged this · 7 years ago
mojomatao reblogged this · 7 years ago -
 goldsakura liked this · 7 years ago
goldsakura liked this · 7 years ago -
 serenehedonist reblogged this · 7 years ago
serenehedonist reblogged this · 7 years ago -
 gabiflower02-blog liked this · 7 years ago
gabiflower02-blog liked this · 7 years ago -
 wowitscorrrin-blog reblogged this · 7 years ago
wowitscorrrin-blog reblogged this · 7 years ago -
 wowitscorrrin-blog liked this · 7 years ago
wowitscorrrin-blog liked this · 7 years ago -
 a2dr3ss8-blog liked this · 7 years ago
a2dr3ss8-blog liked this · 7 years ago -
 qoolqid-blog reblogged this · 7 years ago
qoolqid-blog reblogged this · 7 years ago -
 prodya-blog reblogged this · 7 years ago
prodya-blog reblogged this · 7 years ago -
 knihomol1552-blog liked this · 7 years ago
knihomol1552-blog liked this · 7 years ago -
 katevargas0-blog liked this · 7 years ago
katevargas0-blog liked this · 7 years ago -
 messyboy liked this · 7 years ago
messyboy liked this · 7 years ago -
 cowboydp-blog liked this · 7 years ago
cowboydp-blog liked this · 7 years ago -
 sw33t-stuff liked this · 7 years ago
sw33t-stuff liked this · 7 years ago -
 stardust7635 liked this · 7 years ago
stardust7635 liked this · 7 years ago -
 surprisebear liked this · 7 years ago
surprisebear liked this · 7 years ago -
 alfredocerbino-blog liked this · 7 years ago
alfredocerbino-blog liked this · 7 years ago -
 kryocosmic liked this · 7 years ago
kryocosmic liked this · 7 years ago -
 basswhisperer-blog liked this · 7 years ago
basswhisperer-blog liked this · 7 years ago -
 theemiliodamico liked this · 7 years ago
theemiliodamico liked this · 7 years ago -
 420-itstime-blog liked this · 7 years ago
420-itstime-blog liked this · 7 years ago -
 hypuzzler-blog liked this · 7 years ago
hypuzzler-blog liked this · 7 years ago -
 stephaneraymond-blog liked this · 7 years ago
stephaneraymond-blog liked this · 7 years ago -
 stephaneraymond-blog reblogged this · 7 years ago
stephaneraymond-blog reblogged this · 7 years ago -
 twashings-blog reblogged this · 7 years ago
twashings-blog reblogged this · 7 years ago -
 niworkboo-blog reblogged this · 7 years ago
niworkboo-blog reblogged this · 7 years ago -
 black-rock-shooter-doppelga-blog reblogged this · 7 years ago
black-rock-shooter-doppelga-blog reblogged this · 7 years ago
A pharmacist and a little science sideblog. "Knowledge belongs to humanity, and is the torch which illuminates the world." - Louis Pasteur
215 posts
